from fastMONAI.vision_all import *
from monai.apps import DecathlonDataset
from sklearn.model_selection import train_test_splitPatch-Based Inference
PatchInferenceEngine.
Load data
Load the same dataset and recreate the train/test split from the training notebook.
path = Path('../data')
path.mkdir(exist_ok=True)task = "Task02_Heart"
training_data = DecathlonDataset(root_dir=path, task=task, section="training",
download=True, cache_num=0, num_workers=3)2026-02-03 15:10:15,182 - INFO - Verified 'Task02_Heart.tar', md5: 06ee59366e1e5124267b774dbd654057.
2026-02-03 15:10:15,182 - INFO - File exists: ../data/Task02_Heart.tar, skipped downloading.
2026-02-03 15:10:15,183 - INFO - Non-empty folder exists in ../data/Task02_Heart, skipped extracting.df = pd.DataFrame(training_data.data)
train_df, test_df = train_test_split(df, test_size=0.1, random_state=42)
print(f"Test samples: {len(test_df)}")
test_dfTest samples: 2| image | label | |
|---|---|---|
| 0 | ../data/Task02_Heart/imagesTr/la_030.nii.gz | ../data/Task02_Heart/labelsTr/la_030.nii.gz |
| 1 | ../data/Task02_Heart/imagesTr/la_024.nii.gz | ../data/Task02_Heart/labelsTr/la_024.nii.gz |
Load configuration
Load the patch configuration saved during training. This ensures preprocessing consistency:
- apply_reorder: Whether to reorder to RAS+ orientation
- target_spacing: Target voxel spacing for resampling
- patch_size, patch_overlap, aggregation_mode: Inference parameters
Critical: Mismatched preprocessing parameters will produce incorrect predictions (e.g., mirrored or rotated outputs).
config_dict = load_patch_variables('patch_config.pkl')
print("Loaded configuration:")
for k, v in config_dict.items():
print(f" {k}: {v}")Loaded configuration:
patch_size: [128, 128, 64]
patch_overlap: 0.5
aggregation_mode: hann
apply_reorder: True
target_spacing: [1.25, 1.25, 1.37]
sampler_type: label
label_probabilities: {0: 0.2, 1: 0.8}
samples_per_volume: 8
queue_length: 300
queue_num_workers: 4
keep_largest_component: Truepatch_config = PatchConfig(**config_dict)
apply_reorder = config_dict['apply_reorder']
target_spacing = config_dict['target_spacing']Load trained model
Load the exported learner which contains both the model architecture and best weights.
Two options: 1. Local file: Load best_learner.pkl exported during training 2. MLflow: Download from MLflow artifacts (for experiment tracking workflows)
from fastai.learner import load_learner
import torch
# Option 1: Load from local file
learn = load_learner('models/best_learner.pkl')
# Option 2: Load from MLflow (uncomment to use)
# import mlflow
# run_id = "your_run_id" # Get from MLflow UI
# mlflow.artifacts.download_artifacts(run_id=run_id, artifact_path="model/best_learner.pkl", dst_path="./")
# learn = load_learner('best_learner.pkl')
model = learn.model
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model.to(device)
model.eval()
print(f"Loaded model: {model.__class__.__name__}")
print(f"Device: {device}")Define pre-inference transforms
These transforms MUST match the pre_patch_tfms used during training.
Troubleshooting: If predictions appear mirrored, rotated, or completely wrong, check that
apply_reorder,target_spacing, andpre_inference_tfmsmatch training.
# MUST match training pre_patch_tfms
pre_inference_tfms = [ZNormalization(masking_method='foreground')]Create PatchInferenceEngine
The engine handles the complete sliding-window inference pipeline: 1. Load and preprocess the image (reorder, resample, normalize) 2. Pad if image is smaller than patch size 3. Extract overlapping patches with GridSampler 4. Predict on batches of patches 5. Reconstruct full volume with GridAggregator using Hann windowing
engine = PatchInferenceEngine(
learner=model, # Pass model directly (no Learner needed)
config=patch_config,
pre_inference_tfms=pre_inference_tfms,
batch_size=4
)Single image inference
Use engine.predict() to predict on a single image.
test_path = test_df.iloc[0]['image']
pred, affine = engine.predict(test_path, return_affine=True)
print(f"Input: {test_path}")
print(f"Prediction shape: {pred.shape}")
print(f"Unique values: {torch.unique(pred).tolist()}")Input: ../data/Task02_Heart/imagesTr/la_030.nii.gz
Prediction shape: torch.Size([1, 320, 320, 110])
Unique values: [0, 1]Batch inference
Use patch_inference() to predict on multiple images with optional NIfTI output.
test_paths = test_df['image'].tolist()
predictions = patch_inference(
learner=model, # Pass model directly (no Learner needed)
config=patch_config,
file_paths=test_paths,
pre_inference_tfms=pre_inference_tfms,
save_dir='predictions/patch_heart',
progress=True
)
print(f"\nGenerated {len(predictions)} predictions")
print(f"Saved to: predictions/patch_heart/")
Generated 2 predictions
Saved to: predictions/patch_heart/Evaluate predictions
Evaluate predictions against ground truth using comprehensive metrics.
Metrics: - DSC: Dice score - overlap similarity (higher = better) - HD95: 95th percentile Hausdorff distance in voxels (lower = better) - Sens: Sensitivity - true positive rate (higher = better) - LDR: Lesion detection rate (higher = better) - RVE: Relative volume error (0 = optimal, + = over-seg, - = under-seg)
from fastMONAI.vision_metrics import (calculate_dsc, calculate_haus,
calculate_confusion_metrics,
calculate_lesion_detection_rate,
calculate_signed_rve)
results = []
for i, pred in enumerate(predictions):
img_name = Path(test_df.iloc[i]['image']).name
gt_path = test_df.iloc[i]['label']
# Load ground truth with matching preprocessing
gt = MedMask.create(gt_path, apply_reorder=apply_reorder, target_spacing=target_spacing)
# Prepare 5D tensors [B, C, D, H, W]
pred_5d = pred.unsqueeze(0).float()
gt_5d = gt.data.unsqueeze(0).float()
# Compute metrics using calculate_* functions (for binary masks)
dsc = calculate_dsc(pred_5d, gt_5d).mean().item()
hd95 = calculate_haus(pred_5d, gt_5d).mean().item()
sens = calculate_confusion_metrics(pred_5d, gt_5d, "sensitivity").nanmean().item()
ldr = calculate_lesion_detection_rate(pred_5d, gt_5d).nanmean().item()
rve = calculate_signed_rve(pred_5d, gt_5d).nanmean().item()
results.append({
'Image': img_name, 'DSC': dsc, 'HD95': hd95,
'Sens': sens, 'LDR': ldr, 'RVE': rve
})
results_df = pd.DataFrame(results)
results_dfmonai.metrics.utils get_mask_edges:always_return_as_numpy: Argument `always_return_as_numpy` has been deprecated since version 1.5.0. It will be removed in version 1.7.0. The option is removed and the return type will always be equal to the input type.| Image | DSC | HD95 | Sens | LDR | RVE | |
|---|---|---|---|---|---|---|
| 0 | la_030.nii.gz | 0.938002 | 3.000000 | 0.918056 | 1.0 | -0.042528 |
| 1 | la_024.nii.gz | 0.922835 | 3.316625 | 0.907937 | 1.0 | -0.032287 |
metrics = ['DSC', 'HD95', 'Sens', 'LDR', 'RVE']
print("Test Set Performance Summary")
print("=" * 45)
for metric in metrics:
mean = results_df[metric].mean()
std = results_df[metric].std()
print(f"{metric:8s}: {mean:.4f} ± {std:.4f}")Test Set Performance Summary
=============================================
DSC : 0.9304 ± 0.0107
HD95 : 3.1583 ± 0.2239
Sens : 0.9130 ± 0.0072
LDR : 1.0000 ± 0.0000
RVE : -0.0374 ± 0.0072Visualize predictions
from fastMONAI.vision_plot import show_segmentation_comparison
# Visualize first test case
idx = 0
test_img = MedImage.create(
test_df.iloc[idx]['image'],
apply_reorder=apply_reorder,
target_spacing=target_spacing
)
test_gt = MedMask.create(
test_df.iloc[idx]['label'],
apply_reorder=apply_reorder,
target_spacing=target_spacing
)
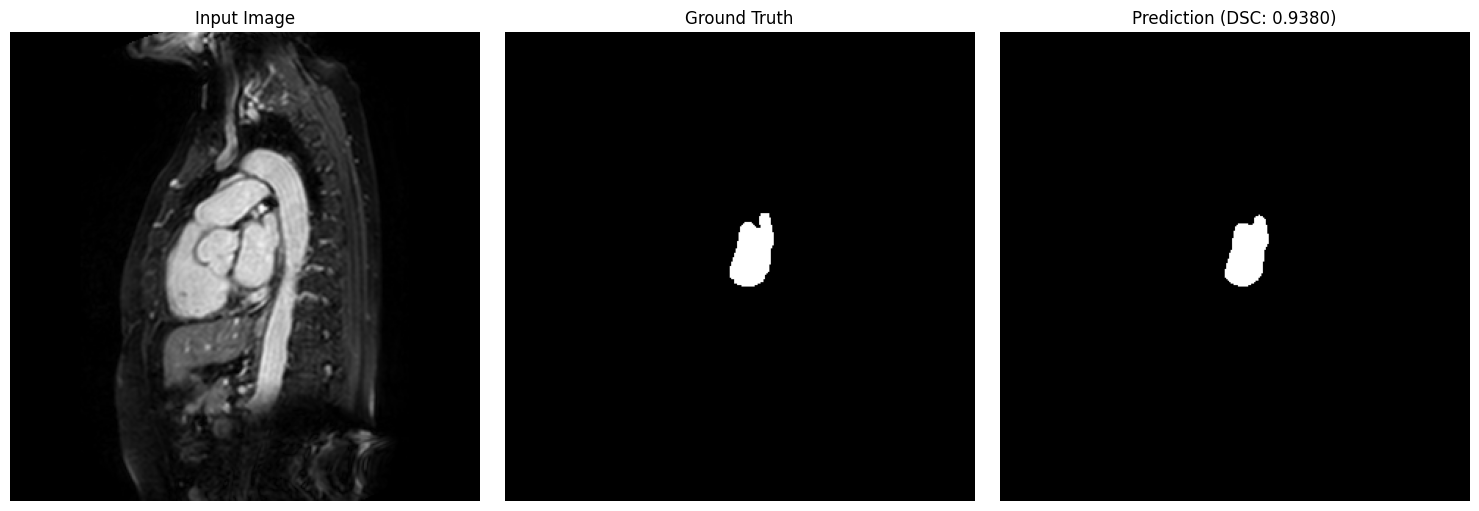
show_segmentation_comparison(
image=test_img,
ground_truth=test_gt,
prediction=predictions[idx],
metric_value=results_df.iloc[idx]['DSC'],
voxel_size=target_spacing,
anatomical_plane=2 # axial view
)
Summary
In this tutorial, we demonstrated patch-based inference for 3D medical image segmentation:
load_patch_variables(): Load configuration saved during training- Direct weight loading: Use
torch.load()to load weights directly (no Learner/DataLoaders needed) PatchInferenceEngine: Sliding-window inference with GridSampler and GridAggregatorpatch_inference(): Batch inference with optional NIfTI output
Key points: - Preprocessing parameters (apply_reorder, target_spacing, pre_inference_tfms) MUST match training - Pass model directly to PatchInferenceEngine - no need for fastai Learner at inference time - Hann windowing (aggregation_mode='hann') produces smooth boundaries between patches - keep_largest_component=True can clean up small spurious predictions
When to use patch-based inference: - Large images that don’t fit in GPU memory - Variable-sized inputs (no resizing needed) - Memory-constrained environments
Tradeoffs: - Slower than full-image inference (multiple forward passes) - Overlap and aggregation add computational overhead - But enables processing of arbitrarily large volumes