from fastMONAI.vision_all import *
from monai.apps import DecathlonDataset
from sklearn.model_selection import train_test_splitPatch-Based Binary Segmentation
MedPatchDataLoaders with lazy loading - volumes are loaded on-demand, keeping memory usage constant regardless of dataset size.
Download external data
We use the MONAI function DecathlonDataset to download the Heart MRI dataset from the Medical Segmentation Decathlon challenge.
path = Path('../data')
path.mkdir(exist_ok=True)task = "Task02_Heart"
training_data = DecathlonDataset(root_dir=path, task=task, section="training",
download=True, cache_num=0, num_workers=3)2026-02-16 12:51:16,418 - INFO - Verified 'Task02_Heart.tar', md5: 06ee59366e1e5124267b774dbd654057.
2026-02-16 12:51:16,418 - INFO - File exists: ../data/Task02_Heart.tar, skipped downloading.
2026-02-16 12:51:16,419 - INFO - Non-empty folder exists in ../data/Task02_Heart, skipped extracting.df = pd.DataFrame(training_data.data)
df.shape(16, 2)Split the labeled data into training and test sets.
train_df, test_df = train_test_split(df, test_size=0.1, random_state=42)
train_df.shape, test_df.shape((14, 2), (2, 2))Analyze training data
Use MedDataset to analyze the dataset and get preprocessing recommendations.
med_dataset = MedDataset(img_list=train_df.label.tolist(), dtype=MedMask, max_workers=12)MedDataset cache: 14 cached, 0 processedmed_dataset.df.head()| path | content_hash | dim_0 | dim_1 | dim_2 | voxel_0 | voxel_1 | voxel_2 | orientation | label_1_volume_mm3 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ../data/Task02_Heart/labelsTr/la_023.nii.gz | a4b0e976cf03b0f1ffdbf79ae91281c4 | 320 | 320 | 110 | 1.25 | 1.25 | 1.37 | RAS+ | 92483.5628 |
| 1 | ../data/Task02_Heart/labelsTr/la_004.nii.gz | db5dcb7cce258526726054f56bf015ef | 320 | 320 | 110 | 1.25 | 1.25 | 1.37 | RAS+ | 125173.0473 |
| 2 | ../data/Task02_Heart/labelsTr/la_007.nii.gz | 4b1c1bf9d0d1adfb12d287c7aa200edc | 320 | 320 | 130 | 1.25 | 1.25 | 1.37 | RAS+ | 118684.8129 |
| 3 | ../data/Task02_Heart/labelsTr/la_022.nii.gz | f9c20e542980932d2ea19a56f3bc7a60 | 320 | 320 | 110 | 1.25 | 1.25 | 1.37 | RAS+ | 71820.1096 |
| 4 | ../data/Task02_Heart/labelsTr/la_011.nii.gz | f1dd0a596e4daf898c3287441971cc06 | 320 | 320 | 120 | 1.25 | 1.25 | 1.37 | RAS+ | 125130.2348 |
data_info_df = med_dataset.summary()suggestion = med_dataset.get_suggestion()
target_spacing = suggestion['target_spacing']
target_spacing[1.25, 1.25, 1.37]stats = med_dataset.get_size_statistics(target_spacing=target_spacing)
print(f"Image sizes (after resampling to {target_spacing}):")
print(f" Min: {stats['min']}")
print(f" Median: {stats['median']}")
print(f" Max: {stats['max']}")
suggested_size = suggest_patch_size(med_dataset, target_spacing=target_spacing)
print(f"\nSuggested patch size: {suggested_size}")Image sizes (after resampling to [1.25, 1.25, 1.37]):
Min: [320.0, 320.0, 90.0]
Median: [320.0, 320.0, 110.0]
Max: [320.0, 320.0, 130.0]
Suggested patch size: [256, 256, 80]Dim 2: patch_size reduced from 96 to 80 to fit smallest volume (min=90, median=110).Configure patch-based training
PatchConfig centralizes all patch-related parameters:
- patch_size: Size of extracted patches
[x, y, z]- should be divisible by 16 for UNet compatibility - samples_per_volume: Number of patches extracted per volume per epoch
- sampler_type:
'uniform'(random) or'label'(foreground-weighted) - label_probabilities: For
'label'sampler, probability of sampling each class - queue_length: Number of patches to keep in memory buffer
- patch_overlap: Overlap for inference (float 0-1 for fraction, or int for pixels)
- aggregation_mode: How to combine overlapping patches (
'hann'for smooth boundaries) - padding_mode: Padding mode when image < patch_size (0 = zero padding, nnU-Net standard)
patch_config = PatchConfig(
patch_size=[160, 160, 80],
samples_per_volume=8,
sampler_type='label',
label_probabilities={0: 0.5, 1: 0.5},
patch_overlap=0.5,
keep_largest_component=True,
target_spacing=target_spacing,
aggregation_mode='hann'
# apply_reorder defaults to True (the common case)
)
print(f"Patch config: {patch_config}")Patch config: PatchConfig(patch_size=[160, 160, 80], patch_overlap=0.5, samples_per_volume=8, sampler_type='label', label_probabilities={0: 0.5, 1: 0.5}, queue_length=300, queue_num_workers=4, aggregation_mode='hann', apply_reorder=True, target_spacing=[1.25, 1.25, 1.37], padding_mode=0, keep_largest_component=True)Alternative: Use
PatchConfig.from_dataset(med_dataset)to auto-configure patch size based on dataset analysis.
Define transforms
Patch-based training uses two stages of transforms:
- pre_patch_tfms: Applied to full volumes before patch extraction (e.g., normalization)
- patch_tfms: Applied to extracted patches during training (augmentations)
Note:
suggest_patch_sizeensures the recommended patch size fits all volumes in the dataset. No automatic padding is applied during training.
Critical:
pre_patch_tfmsmust match between training and inference for consistent preprocessing.
# Pre-patch transforms (applied to full volumes by Queue workers)
pre_patch_tfms = [ZNormalization(masking_method='foreground')]
patch_tfms = [
RandomAffine(scales=(0.7, 1.4), degrees=30, translation=(25, 25, 10), p=0.2),
RandomAnisotropy(downsampling=(1.5, 3), p=0.25),
RandomGamma(log_gamma=(-0.3, 0.3), p=0.3),
RandomIntensityScale(scale_range=(0.75, 1.25), p=0.1),
RandomNoise(std=0.1, p=0.1),
RandomBlur(std=(0.5, 1.0), p=0.2),
RandomFlip(axes='LRAPIS', p=0.5),
]Alternative: GPU-batched augmentation
For long training runs (e.g., hundreds of epochs), GPU-batched augmentation can significantly reduce training time by moving transforms from CPU to GPU. Instead of per-sample TorchIO transforms (patch_tfms), gpu_patch_augmentations creates a batched augmentation pipeline that operates on GPU tensors directly.
Use gpu_augmentation instead of patch_tfms (they are mutually exclusive).
# # GPU augmentation alternative (uncomment to use instead of patch_tfms above)
# gpu_aug = gpu_patch_augmentations(patch_config.patch_size, patch_config.target_spacing)
#
# # Then pass gpu_augmentation instead of patch_tfms to from_df:
# # dls = MedPatchDataLoaders.from_df(
# # ..., gpu_augmentation=gpu_aug, ... # replaces patch_tfms=patch_tfms
# # )Create patch-based DataLoaders
MedPatchDataLoaders uses lazy loading: - Only file paths are stored at creation time (~0 MB) - Volumes are loaded on-demand by Queue workers
bs = 4
dls = MedPatchDataLoaders.from_df(
df=train_df,
img_col='image',
mask_col='label',
valid_pct=0.1,
patch_config=patch_config,
pre_patch_tfms=pre_patch_tfms,
patch_tfms=patch_tfms,
bs=bs,
seed=42
)
print(f"Training subjects: {len(dls.train.subjects_dataset)}")
print(f"Validation subjects: {len(dls.valid.subjects_dataset)}")Training subjects: 13
Validation subjects: 1# Visualize a batch of patches
batch = next(iter(dls.train))
x, y = batch
print(f"Batch shape - Image: {x.shape}, Mask: {y.shape}")Batch shape - Image: torch.Size([4, 1, 160, 160, 80]), Mask: torch.Size([4, 1, 160, 160, 80])dls.show_batch(anatomical_plane=2, max_n=2, overlay=False)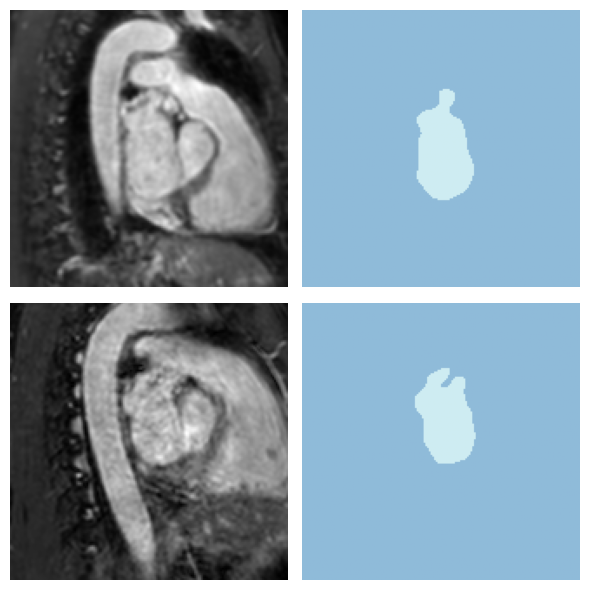
Create and train a 3D model
We use MONAI’s UNet with: - out_channels=2: Softmax output (background + foreground) - Instance normalization: Common in medical imaging - DiceCELoss: Combines Dice loss with Cross-Entropy for stable training
from monai.networks.nets import UNet
from monai.networks.layers import Norm
from monai.losses import DiceCELoss- DiceCELoss: Combines Dice loss with Cross-Entropy for stable training.
batch=Truecomputes Dice by pooling TP/FP/FN across all samples in the batch before computing the ratio (nnU-Net default), rather than averaging per-sample Dice scores. This provides more stable gradients when some patches contain little or no foreground.
model = UNet(
spatial_dims=3,
in_channels=1,
out_channels=2,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2,
norm=Norm.INSTANCE
)
loss_func = CustomLoss(loss_func=DiceCELoss(
to_onehot_y=True,
softmax=True,
include_background=False,
batch=True
))We use AccumulatedDice metric which accumulates true positives, false positives, and false negatives across all validation batches before computing Dice.
learn = Learner(dls, model, loss_func=loss_func, metrics=[AccumulatedDice(n_classes=2)])learn.lr_find()SuggestedLRs(valley=0.0012022644514217973)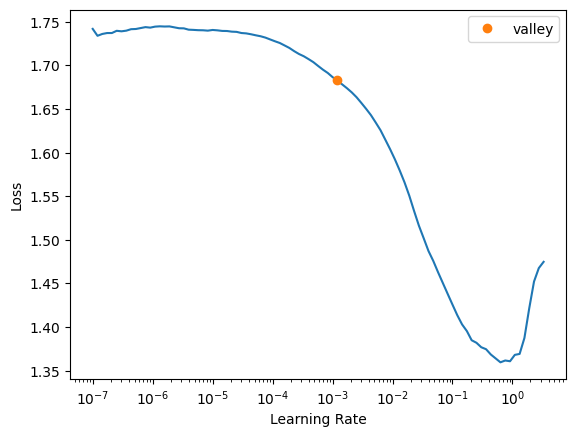
lr = 1e-3best_model_fname = "best_heart_patch"
save_best = EMACheckpoint(
monitor='accumulated_dice',
momentum=0.9,
comp=np.greater,
fname=best_model_fname,
with_opt=False
)# All params (size, transforms, loss, model name, etc.) are extracted from learn automatically
mlflow_callback = create_mlflow_callback(learn, experiment_name="Task02_Heart_Patch", dataset_version=med_dataset.fingerprint)
learn.fit_one_cycle(40, lr, cbs=[mlflow_callback, save_best])2026/02/16 12:51:56 INFO alembic.runtime.migration: Context impl SQLiteImpl.
2026/02/16 12:51:56 INFO alembic.runtime.migration: Will assume non-transactional DDL.Logged train/val split (14 rows) to MLflow artifacts| epoch | train_loss | valid_loss | accumulated_dice | time |
|---|---|---|---|---|
| 0 | 1.674814 | 1.649808 | 0.049942 | 00:10 |
| 1 | 1.622354 | 1.583473 | 0.070154 | 00:12 |
| 2 | 1.571586 | 1.512572 | 0.093310 | 00:11 |
| 3 | 1.508447 | 1.407170 | 0.151716 | 00:11 |
| 4 | 1.423123 | 1.286892 | 0.190416 | 00:11 |
| 5 | 1.312932 | 1.126350 | 0.286872 | 00:11 |
| 6 | 1.192703 | 1.050131 | 0.222503 | 00:12 |
| 7 | 1.067632 | 0.993860 | 0.202609 | 00:11 |
| 8 | 0.955299 | 0.838112 | 0.346686 | 00:11 |
| 9 | 0.858103 | 0.907975 | 0.220042 | 00:11 |
| 10 | 0.786617 | 0.799449 | 0.328615 | 00:11 |
| 11 | 0.699878 | 0.636010 | 0.484181 | 00:11 |
| 12 | 0.591402 | 0.482632 | 0.627061 | 00:12 |
| 13 | 0.520575 | 0.405796 | 0.682028 | 00:12 |
| 14 | 0.492879 | 0.629759 | 0.469315 | 00:11 |
| 15 | 0.460075 | 0.452520 | 0.628223 | 00:11 |
| 16 | 0.403452 | 0.546032 | 0.537498 | 00:11 |
| 17 | 0.365366 | 0.481454 | 0.591646 | 00:11 |
| 18 | 0.328365 | 0.440150 | 0.627600 | 00:11 |
| 19 | 0.296089 | 0.338266 | 0.716846 | 00:11 |
| 20 | 0.261054 | 0.382205 | 0.682141 | 00:11 |
| 21 | 0.251172 | 0.524608 | 0.535143 | 00:13 |
| 22 | 0.246274 | 0.478194 | 0.595964 | 00:11 |
| 23 | 0.231552 | 0.357881 | 0.699248 | 00:12 |
| 24 | 0.224238 | 0.384899 | 0.677488 | 00:12 |
| 25 | 0.223415 | 0.374642 | 0.683922 | 00:11 |
| 26 | 0.221865 | 0.388269 | 0.673840 | 00:11 |
| 27 | 0.216021 | 0.362696 | 0.694873 | 00:11 |
| 28 | 0.202212 | 0.267767 | 0.776801 | 00:11 |
| 29 | 0.192795 | 0.324323 | 0.724529 | 00:11 |
| 30 | 0.185226 | 0.286899 | 0.759452 | 00:11 |
| 31 | 0.176141 | 0.286998 | 0.753827 | 00:11 |
| 32 | 0.172152 | 0.303943 | 0.743113 | 00:11 |
| 33 | 0.164531 | 0.261736 | 0.778175 | 00:11 |
| 34 | 0.161059 | 0.294292 | 0.749676 | 00:11 |
| 35 | 0.157056 | 0.303720 | 0.743407 | 00:11 |
| 36 | 0.160109 | 0.302797 | 0.741491 | 00:12 |
| 37 | 0.158391 | 0.273787 | 0.768147 | 00:11 |
| 38 | 0.156991 | 0.313097 | 0.740300 | 00:12 |
| 39 | 0.152525 | 0.274406 | 0.769968 | 00:12 |
Training finished. Logging model artifacts to MLflow...
Logged best model weights: best_weights.pth
Logged final epoch learner: final_learner.pkl
Loaded best model weights (best_heart_patch) for best learner export
Logged best epoch learner: best_learner.pklSaved file doesn't contain an optimizer state.
2026/02/16 12:59:47 WARNING mlflow.pytorch: Saving pytorch model by Pickle or CloudPickle format requires exercising caution because these formats rely on Python's object serialization mechanism, which can execute arbitrary code during deserialization.The recommended safe alternative is to set 'export_model' to True to save the pytorch model using the safe graph model format.MLflow run completed. Run ID: 6b3655d7caeb49ceb0be7dfbfd7cbfd1Registered model 'UNet' already exists. Creating a new version of this model...
Created version '25' of model 'UNet'.learn.recorder.plot_loss();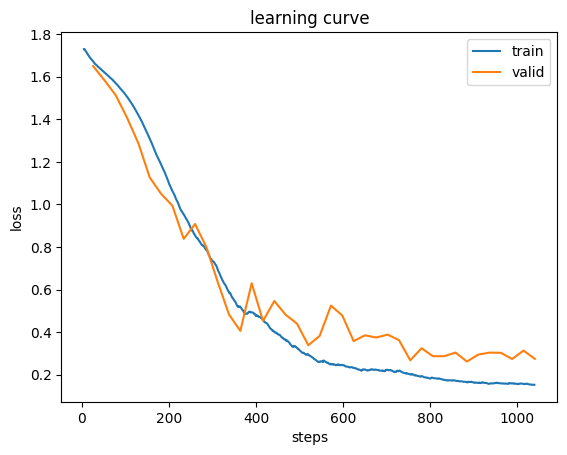
Save configuration for inference
The following parameters MUST be identical between training and inference: - apply_reorder: Whether to reorder to RAS+ orientation - target_spacing: Target voxel spacing for resampling - pre_patch_tfms: Preprocessing transforms (e.g., ZNormalization)
Save these to a pickle file for the inference notebook to load.
store_patch_variables(
pkl_fn='patch_config.pkl',
patch_size=patch_config.patch_size,
patch_overlap=patch_config.patch_overlap,
aggregation_mode=patch_config.aggregation_mode,
apply_reorder=patch_config.apply_reorder,
target_spacing=patch_config.target_spacing,
sampler_type=patch_config.sampler_type,
label_probabilities=patch_config.label_probabilities,
samples_per_volume=patch_config.samples_per_volume,
queue_length=patch_config.queue_length,
queue_num_workers=patch_config.queue_num_workers,
keep_largest_component=patch_config.keep_largest_component
)
print("Configuration saved to patch_config.pkl")Configuration saved to patch_config.pklEvaluate on validation set
Evaluate the trained model on the validation set using sliding-window patch inference.
Metrics: - DSC: Dice score - overlap similarity (higher = better) - HD95: 95th percentile Hausdorff distance in voxels (lower = better) - Sens: Sensitivity - true positive rate (higher = better) - LDR: Lesion detection rate (higher = better) - RVE: Relative volume error (0 = optimal, + = over-seg, - = under-seg)
# Extract file paths from validation subjects
val_subjects = dls.valid.subjects_dataset
val_img_paths = [str(s['image'].path) for s in val_subjects]
val_mask_paths = [str(s['mask'].path) for s in val_subjects]
print(f"Validation set: {len(val_img_paths)} images")Validation set: 1 images# Load the best model (saved by EMACheckpoint based on smoothed accumulated_dice)
learn.load(best_model_fname)
print(f"Loaded best model: {best_model_fname}")
learn.export(f"models/best_learner.pkl")
save_dir = Path('predictions/patch_heart_val')
predictions = patch_inference(
learner=learn,
config=patch_config,
file_paths=val_img_paths,
pre_inference_tfms=pre_patch_tfms,
save_dir=str(save_dir),
progress=True,
tta=True
)
print(f"\nSaved {len(predictions)} predictions to {save_dir}/")Loaded best model: best_heart_patchSaved file doesn't contain an optimizer state.
Saved 1 predictions to predictions/patch_heart_val/from fastMONAI.vision_metrics import (calculate_dsc, calculate_haus,
calculate_confusion_metrics,
calculate_lesion_detection_rate,
calculate_signed_rve)
results = []
for i, pred in enumerate(predictions):
img_name = Path(val_img_paths[i]).name
gt_path = val_mask_paths[i]
# Load ground truth with matching preprocessing
gt = MedMask.create(
gt_path,
apply_reorder=patch_config.apply_reorder,
target_spacing=patch_config.target_spacing
)
# Prepare 5D tensors [B, C, D, H, W]
pred_5d = pred.unsqueeze(0).float()
gt_5d = gt.data.unsqueeze(0).float()
# Compute metrics using calculate_* functions (for binary masks)
dsc = calculate_dsc(pred_5d, gt_5d).mean().item()
hd95 = calculate_haus(pred_5d, gt_5d).mean().item()
sens = calculate_confusion_metrics(pred_5d, gt_5d, "sensitivity").nanmean().item()
ldr = calculate_lesion_detection_rate(pred_5d, gt_5d).nanmean().item()
rve = calculate_signed_rve(pred_5d, gt_5d).nanmean().item()
results.append({
'image': img_name, 'dsc': dsc, 'hd95': hd95,
'sensitivity': sens, 'ldr': ldr, 'rve': rve
})
results_df = pd.DataFrame(results)monai.metrics.utils get_mask_edges:always_return_as_numpy: Argument `always_return_as_numpy` has been deprecated since version 1.5.0. It will be removed in version 1.7.0. The option is removed and the return type will always be equal to the input type.# Log validation metrics to MLflow (metrics auto-inferred from numeric columns)
mlflow_callback.log_metrics_table(results_df, display=True)
mlflow_callback.log_metrics(
{f'val_{m}': results_df[m].mean() for m in results_df.select_dtypes(include='number').columns}
)
mlflow_callback.log_dataframe(results_df)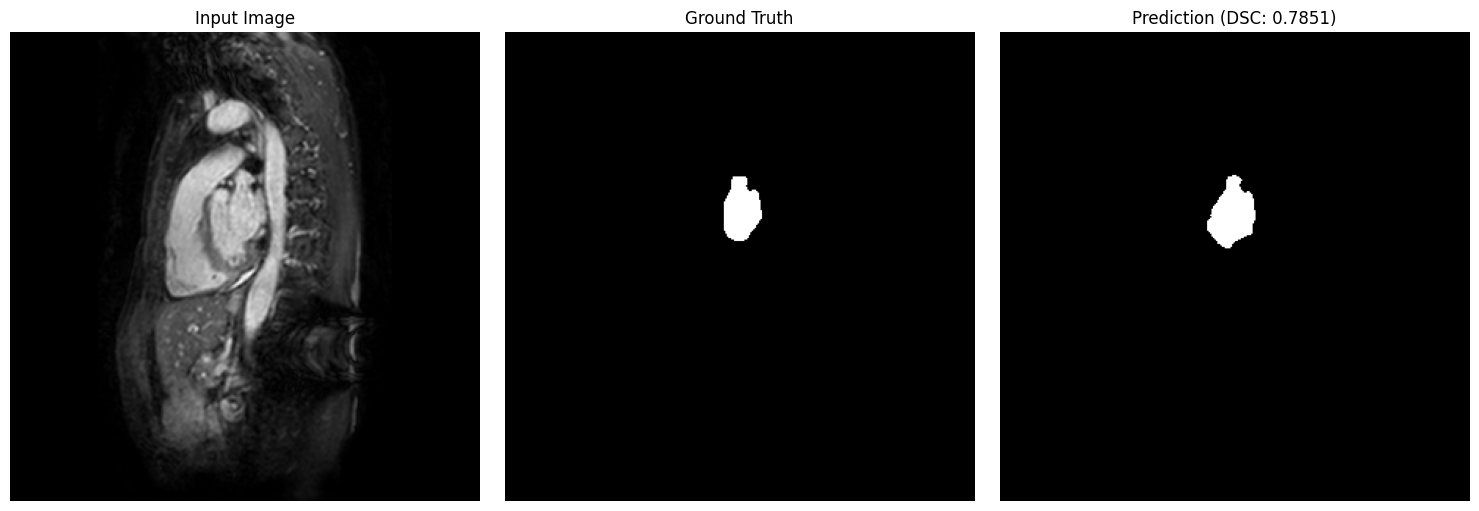
Logged metrics table to MLflow run 6b3655d7caeb49ceb0be7dfbfd7cbfd1
Logged 5 metric(s) to MLflow run 6b3655d7caeb49ceb0be7dfbfd7cbfd1
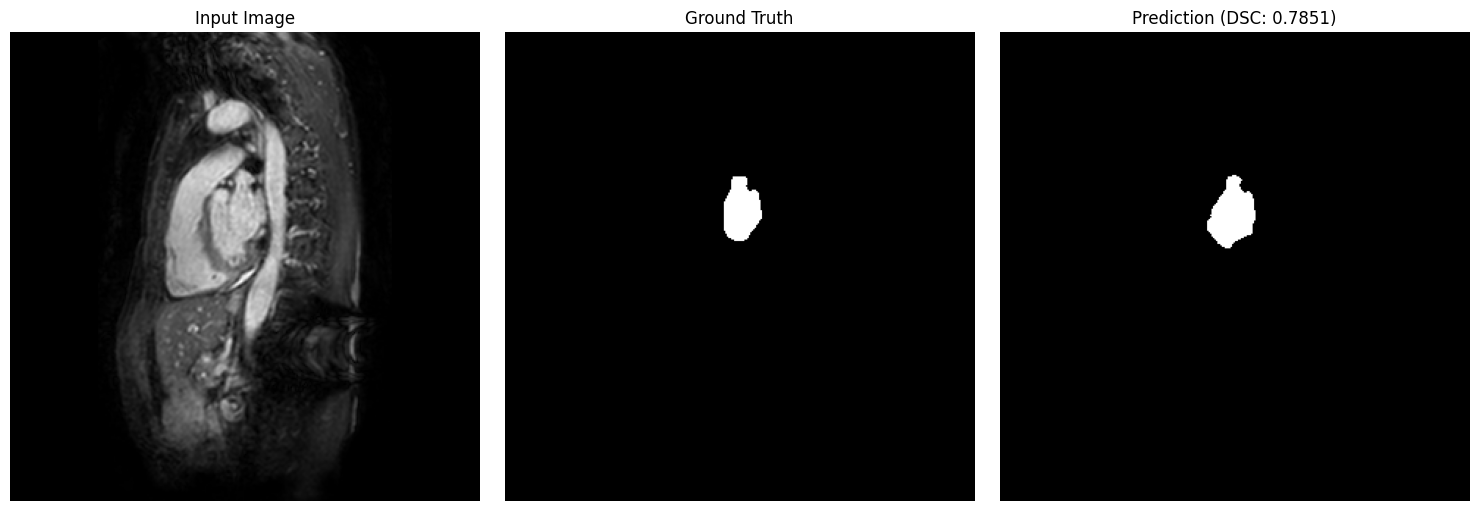
Logged DataFrame (1 rows) to MLflow run 6b3655d7caeb49ceb0be7dfbfd7cbfd1from fastMONAI.vision_plot import show_segmentation_comparison
idx = 0
val_img = MedImage.create(
val_img_paths[idx],
apply_reorder=patch_config.apply_reorder,
target_spacing=patch_config.target_spacing
)
val_gt = MedMask.create(
val_mask_paths[idx],
apply_reorder=patch_config.apply_reorder,
target_spacing=patch_config.target_spacing
)
show_segmentation_comparison(
image=val_img,
ground_truth=val_gt,
prediction=predictions[idx],
metric_value=results_df.iloc[idx]['dsc'],
voxel_size=patch_config.target_spacing,
anatomical_plane=2 # axial view
)
View experiment tracking
mlflow_ui = MLflowUIManager()
mlflow_ui.start_ui()TrueSummary
In this tutorial, we demonstrated patch-based training and evaluation for 3D medical image segmentation:
Training: 1. PatchConfig: Centralized configuration for patch size, sampling, and inference parameters 2. MedPatchDataLoaders: Memory-efficient lazy loading with TorchIO Queue 3. Two-stage transforms: pre_patch_tfms (full volume) + patch_tfms (training augmentation) 4. AccumulatedDice: nnU-Net-style accumulated metric for reliable validation 5. store_patch_variables(): Save configuration for inference consistency
Evaluation: 6. patch_inference(): Batch sliding-window inference with NIfTI output 7. Comprehensive metrics: DSC, HD95, Sensitivity, LDR, RVE
Key takeaway: Preprocessing parameters (apply_reorder, target_spacing, pre_inference_tfms) MUST match between training and inference for correct predictions.
mlflow_ui.stop()